No organelles? No problem! WSU lab discovery shows how bacteria get the job done
A lot more goes on inside prokaryotic cells than once thought, according to research in a WSU lab. While they may not have the neat, membrane-bound organelles of eukaryotic cells to handle their inner workings, prokaryotic cells are able to survive and function by making their own membrane-free, blob-like versions of organelles when needed.
Eukaryotes also have some of these nonmembrane-bound structures, such as the RNA factory called the nucleolus, but scientists lumped most of them under the generic name of granules or bodies and gave them little attention ... until recently. "There has been a revolution in the past decade, where people have started using more modern technology to look at these structures in eukaryotic cells and begin understanding more about them," said Jared Schrader, Ph.D., WSU associate professor of biological sciences. He and his research group joined that revolution by asking whether prokaryotic cells also make similar structures, and they quickly had an answer: Yes.
In 2018, Schrader and his group published their discovery of a bacterial version of the eukaryotic structures known as p-bodies and stress granules. These membrane-less eukaryotic structures are involved in degrading RNA, an important step in controlling gene activity and maintaining proper cell function. The bacterial version, which they named bacterial ribonucleoprotein bodies, or BR-bodies for short, also had the job of degrading RNA to keep cells operating smoothly.
With just a small protein and often some nucleic acids, prokaryotic cells use this simple phase transition to generate a structure, and this structure is able to organize key biochemical pathways.

Beyond identifying BR-bodies, Schrader's group went further and investigated how these membrane-free blobs formed. "We found that we could just put their component molecules (mainly proteins and RNA) in a test tube, and they would form droplets on their own. Nothing else was required. It was pure self-assembly," he described. This self-assembly was particularly interesting because it relied on a mechanism somewhat analogous to what happens in well-shaken vinaigrette salad dressing. If left alone, the watery vinegar and the oil will soon begin to separate from one another because their molecules have different properties, he explained.
The same thing happens in BR bodies, but the separation happens a bit differently. "Certain regions of the proteins in BR bodies favor sticking to each other and separating from the other molecules in the cells, and that is what allows them to create distinct droplets," Schrader said. Using a microscope, he and his research team were able to actually watch small droplets begin to form and then fuse together to create bigger and bigger blobs.
This mechanism of membrane-free blob creation is called liquid-liquid phase separation. "The cool thing about BR-body formation in prokaryotes is that you don't need the huge set of enzymes, lipids and proteins that eukaryotic cells need to make functional membrane-bound organelles," he remarked. "With just a small protein and often some nucleic acids, prokaryotic cells use this simple phase transition to generate a structure, and this structure is able to organize key biochemical pathways."
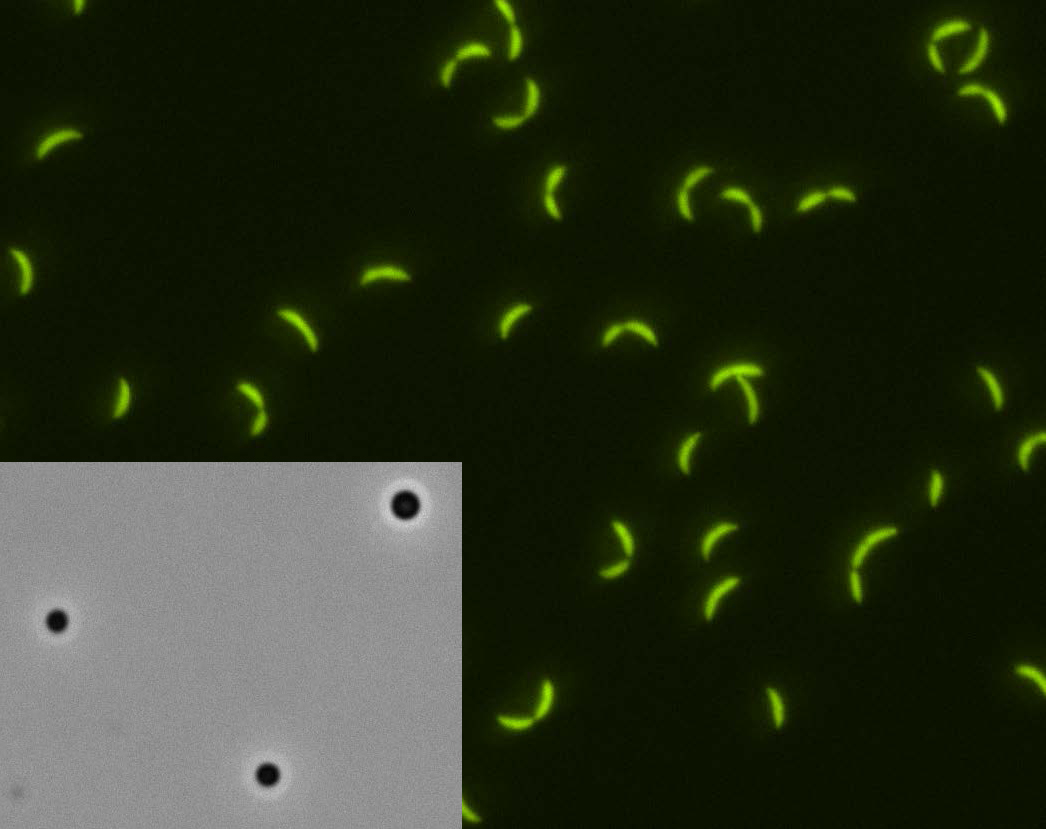
In addition to being the first to report this liquid-liquid phase separation in prokaryotic cells, Schrader and his group also noted that the droplets had binding sites that would target and acquire those enzymes necessary to degrade RNA. "Since then, a lot of other labs have found many other bacterial structures that also can assemble into 'biomolecular condensates,' which is the new, formal name for these non-membrane bound structures in cells," he said.
The WSU research group is now tinkering with the BR-body enzymes to learn which enzymes slow down or speed up RNA degradation, and exactly how they do it. "RNA degradation has many steps involved in it, and we're finding that some specific steps get messed up if a particular enzyme is missing. As an example, a step may not happen as fast, which causes the next step in the process to also slow, which can then disrupt the order of events so nothing is happening in the right place and time," he said.
In addition, Schrader's group has branched out from its original study species, a freshwater bacterium (Caulobacter crescentus), and is seeing similar biomolecular condensates and self-assembly processes in about a dozen other species representing a diversity of bacteria. "We're trying to get the foundational knowledge to determine if these blobs and their self-assembly is universal across all bacteria, including bacterial pathogens," he said.
Such a broader understanding could lead to a wide variety of applications, according to Schrader. "It could perhaps reveal new targets for next-generation antibiotics, or make it possible to engineer blobs that can help speed up the biofermentation of products like biofuels, or to generate all sorts of other useful bioproducts."
These future applications are a very exciting part of this work, but to get to that point, researchers have to do the groundwork, he noted. "And that's what we're doing in my lab. We're getting a basic idea of how these biomolecular condensates work in the first place, and how many bacterial cells have them."
It could perhaps reveal new targets for next-generation antibiotics, or make it possible to engineer blobs that can help speed up the biofermentation of products like biofuels or to generate all sorts of other useful bioproducts.
More information about the Schrader group's discovery of BR-bodies and their self-assembly, including the liquid-liquid phase separation, is available at:
