Hiding in plain sight: Generic protein could be key to fighting dangerous viral infections
Breakthrough work in the lab of Athar Ansari is revealing that a rather small and mostly ignored protein in our cells — a protein called a general transcription factor — likely plays quite a big part in allowing dangerous viruses to infect us.

In recognition of the potential significance of this research, his WSU lab received a $1.05 million, five-year grant from the National Institute of Health to understand the details.
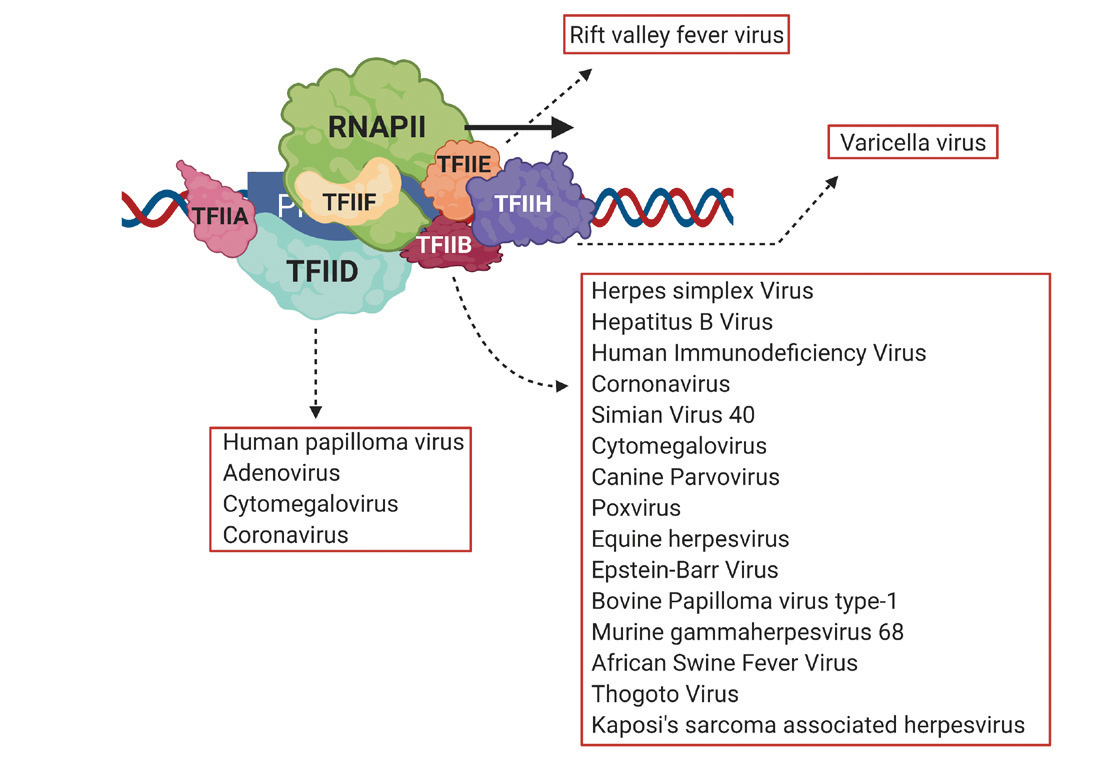
Studies over the past decade have mentioned that one virus or another seems to target the same protein molecule, known as transcription factor IIB (TFIIB). It wasn’t until 2021, however, that Ansari and his research group connected the dots and realized that TFIIB could be a key to fighting a wide range of dangerous viruses, including those responsible for COVID-19, hepatitis B and AIDS.
Ansari actually started studying TFIIB as far back as the mid-1990s, when he was a postdoctoral researcher looking into DNA transcription or the way each of the genes – specific stretches of the DNA code – is read. In transcription, the enzyme RNA polymerase reads the gene from one end to the other, and a handful of transcription factors, including TFIIB, direct the RNA polymerase to just the right initiation point in the DNA code, known as the promoter region, so it can begin reading the targeted gene.
For many years, scientists thought RNA polymerase continued reading a gene straight across until it reached a snippet of DNA code that told it to stop or terminate and then the RNA polymerase assumedly fell away after making that one pass. In 2005, however, Ansari was part of a research group (led by Michael Hampsey, Ph.D., University of Medicine and Dentistry of New Jersey) that found more to the story: somehow, the termination and promoter regions for a gene could physically link up, which caused the gene to form a loop. As a result, the RNA polymerase would finish reading the gene and be back at the initiation point, so it could immediately begin reading the gene again.
Once Ansari joined Wayne State in 2006 and started his own lab, he and his group continued unraveling the mystery of gene looping and subsequently discovered that TFIIB was a lynchpin in the looping mechanism. “We used a number of independent methods and everything pointed to the same thing: TFIIB interacts with the termination end and that interaction is the basis of gene looping,” Ansari said. “We were not expecting that, but the evidence was conclusive.”
While exploring gene looping, mainly by conducting experiments on budding yeast (a common model for genetics studies), Ansari stayed up to date on other groups’ findings about transcription and began to note scattered references that individual viruses might be targeting TFIIB. “In keeping track of all the new papers, I found it interesting that even among those people who were working on viruses and seeing TFIIB-targeting, Athar Ansari nobody was really paying attention to TFIIB,” he said. His group decided to do that legwork. “We found at least 30 papers that noted TFIIB-targeting and we put all that information together into one review article, which we published in 2021,” he said. “That article put forth the hypothesis that TFIIB is targeted because of this multifunctional aspect and involvement in initiation, termination and gene looping.”
Inspired by that bigger picture, Ansari and his research group have delved deeper and found that TFIIB not only interacts with the termination end of transcription but is actually necessary for terminating the transcription process. “In the budding yeast model, we have shown that at least 73 percent of the genes are dependent on TFIIB for termination of transcription,” he said. A new research article about those results is in the works and to take the work even further, his lab is beginning to investigate the prominence of TFIIB-mediated looping among genes.
Overall, Ansari estimates that at least half of all human pathogenic viruses zero in on TFIIB. They hijack transcription so that the human DNA replication machinery deviates from its job of copying our own genes and instead goes to work making copies upon copies of the virus’s genes. In fact, hijacking transcription is the only way viruses can replicate. If viruses use TFIIB to access transcription, this mechanism might also present an avenue to shut down viral transcription.
In that vein, Ansari’s group is using the new $1.05 million grant from the National Institutes of Health to build up his lab staff, including new postdoctoral research and two more graduate students, and begin a collaboration with a virologist to develop additional models of investigation. The goal, he said, is to understand exactly how a virus usurps TFIIB to take over transcription, which will help virologists gain a much clearer view of the potential vulnerabilities of human cells. Ultimately, these insights have a high potential to lead to new viral therapeutics.
“I am looking forward to this work,” Ansari said, noting that it will be very interesting on a basic science level, but also on a practical, translational level. “If we can figure out the different aspects of the transcription cycle, then we can really play a very important role in understanding pathogenesis in human cells. If we can identify a common factor that many viruses use, we have a much better chance of designing a therapeutic that can target not just one virus, but all of the viruses. That is really important.”